BioWarehouse is a component of the Bio-SPICE project. BioWarehouse is an open-source software environment for integrating a set of biological databases into a single physical database management system for data management, mining, and exploration.
Key features of BioWarehouse:
The BioWarehouse is populated using loader programs that translate the flat file representation of a source database into the warehouse schema. A loader is provided for each source database supported by BioWarehouse. Once loaded within a BioWarehouse instance running on e.g. MySQL, a set of source DBs can now be queried together.
Some loaders are specific to a data format rather than to a single source database. For example, the BioPAX and MAGE-ML loaders can load any database that is in BioPAX or MAGE-ML format, respectively.
BioWarehouse loaders:
|
Source DB |
Contents |
Language |
Citation |
|
BioCyc DBs |
Genomes, genes, proteins, metabolic pathways, reactions,
compounds |
C |
|
| BioPAX format | BioPAX format describes biological pathway and protein interaction data. Currently this loader can process BioPAX Level 2 only -- protein interaction data. | JAVA | |
|
Comprehensive Microbial Resource (CMR) |
Genomes, genes, proteins, reactions |
C |
|
|
ENZYME
DB |
Reactions, proteins |
JAVA |
|
| Eco2dbase | E. coli 2D protein gel database | JAVA | |
|
GenBank – bacteria only |
Bacterial genes and proteins |
JAVA |
|
|
A controlled vocabulary to describe gene and gene
product attributes |
JAVA |
|
|
|
|
Genomes, genes, proteins, metabolic pathways, reactions,
compounds |
C |
|
| MetaCyc Ontology | The MetaCyc ontology of metabolic pathways, and
the MetaCyc ontology of chemical compounds |
C | |
| MAGE-ML format | The MAGE-ML file format describes gene expression datasets | JAVA | |
|
Taxonomical organism classification |
C |
||
|
UniProt (Swiss-Prot
and TrEMBL) |
Protein knowledgebase |
JAVA |
The BioWarehouse schema (schema documentation) is designed to capture as much of the data of each component DB as possible within a uniform representation.
For example, in encoding data from a set of source DBs pertaining to proteins, BioWarehouse uses a single set of schema definitions that spans all attributes of proteins found across this set of DBs. This approach eliminates the semantic heterogeneity present in these DBs, allowing users to query all protein sequence DBs using the same schema. Such sharing of tables is applied wherever practical. The translation from the component DB to the warehouse is achieved by the DB loaders, which convert the conceptualization used in each component DB into the conceptualization used by the warehouse schema.
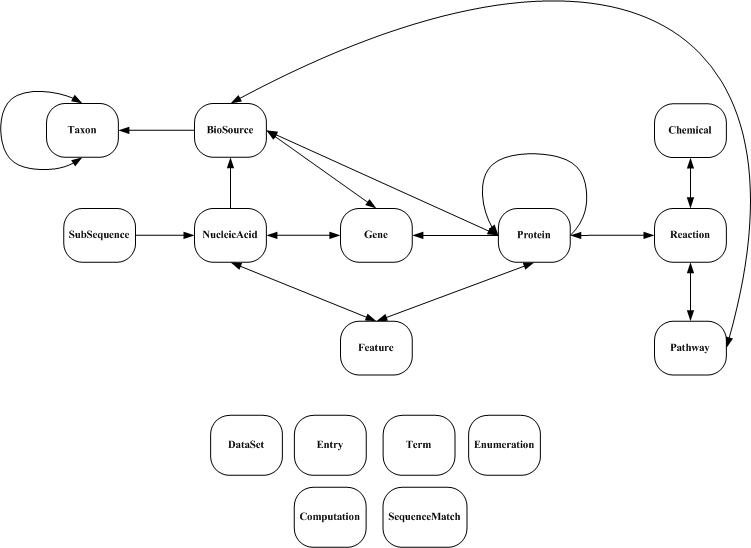
The major biological objects of the BioWarehouse and their interrelationships are depicted below. Arrows indicate that the objects in those tables can refer to entries in the same table.

For each loader, there are two pieces of
documentation:
how to build and run the loader, and a "manual" for developers
describing the details of the loader implementation and schema mappings.
Overall documentation:
All of the overall documentation is listed in a table of contents at doc/index.html. The TOC also has a table listing some statistics about all the loaders (latest supported version of the data, input size, #Objects loader, load time, etc.)
Please contact .
Projects using BioWarehouse include:
Bio-SPICE use
cases
SRI’s Enzyme
Genomics
project
SRI’s Pathway
Hole Filling project
Poster
presented at 2004
Bio-SPICE PI meeting
BioWarehouse paper in BMC Bioinformatics.
The BioWarehouse team:
Peter D. Karp, Principal
Investigator
Thomas J Lee
Yannick Pouliot
Valerie Wagner
Tomer Altman
Nan Guo
David Dunkley
BioWarehouse developed was supported by DARPA contract F30602-01-C-0153, and by NIH grants GM077905 and GM080746.
For support and inquiries please contact .