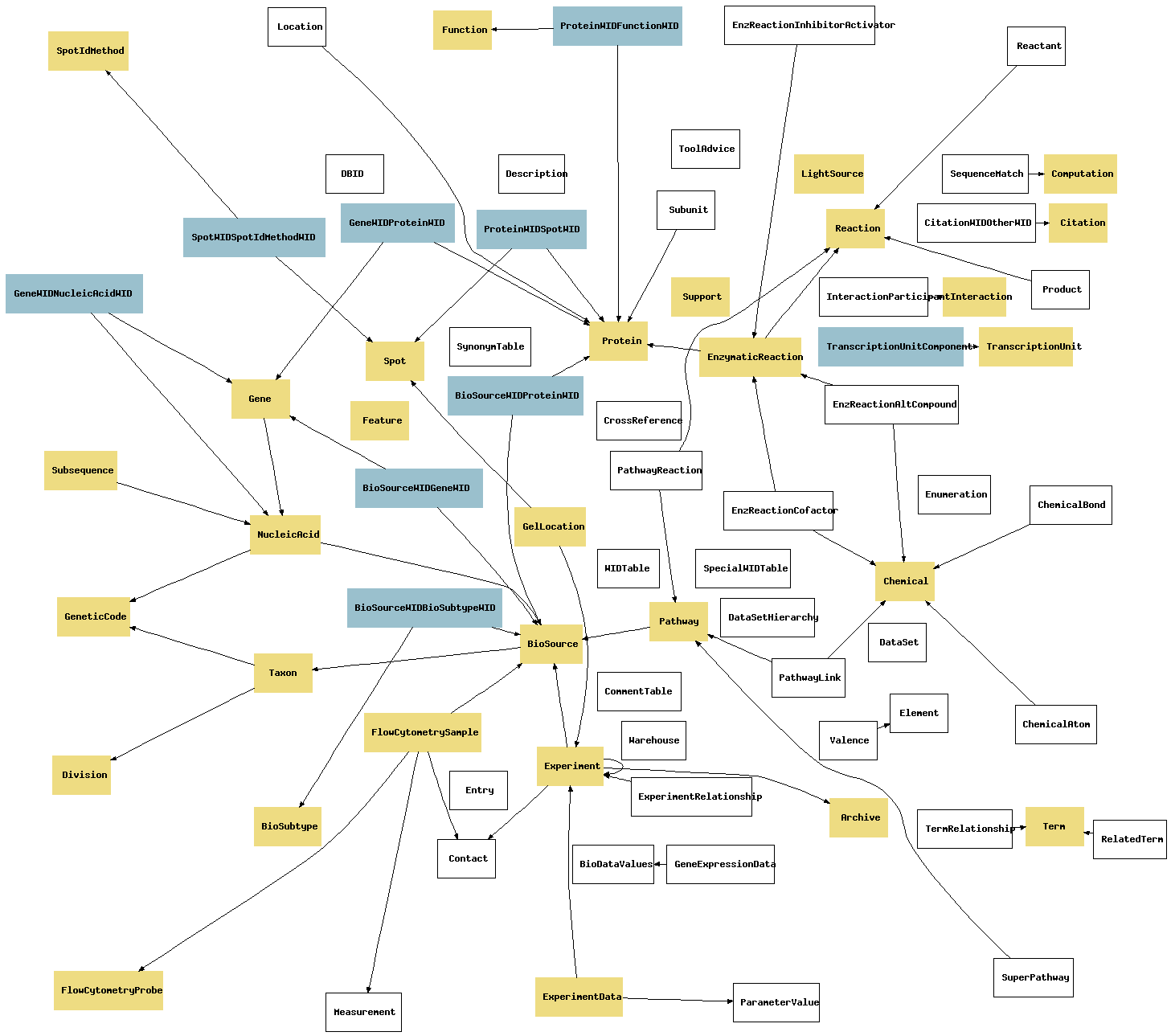
| Table | Comment |
|---|---|
| Archive | Defines an association between a warehouse entity and an external representation or depiction of that entity in a well-defined format. Can be used to store the contents itself, or to indicate that an archive is present at a URL. |
| Array_ | Represents MAGE Class Array The physical substrate along with its features and their annotation |
| ArrayDesign | Represents MAGE Class ArrayDesign Describes the design of an gene expression layout. In some cases this might be virtual and, for instance, represent the output from analysis software at the composite level without reporters or features. Includes MAGE Class PhysicalArrayDesign |
| ArrayDesignWIDCompositeGrpWID | Represents n..n association between ArrayDesign and CompositeGroup The grouping of like CompositeSequence together. If more than one technology type occurs on the array, such as the mixing of Cloned BioMaterial and Oligos, then there would be multiple CompositeGroups to segregate the technology types. |
| ArrayDesignWIDContactWID | Represents n..n association between ArrayDesign and Contact The primary contact for information on the array design |
| ArrayDesignWIDReporterGroupWID | Represents n..n association between ArrayDesign and ReporterGroup The grouping of like Reporter together. If more than one technology type occurs on the array, such as the mixing of Cloned BioMaterial and Oligos, then there would be multiple ReporterGroups to segregate the technology types. |
| ArrayGroup | Represents MAGE Class ArrayGroup An array package is a physical platform that contains one or more arrays that are separately addressable (e.g. several arrays that can be hybridized on a single microscope slide) or a virtual grouping together of arrays. The array package that has been manufactured has information about where certain artifacts about the array are located for scanning and feature extraction purposes. |
| ArrayGroupWIDArrayWID | Represents n..n association between ArrayGroup and Array Association between an ArrayGroup and its Arrays, typically the ArrayGroup will represent a slide and the Arrays will be the manufactured so that they may be hybridized separately on that slide. |
| ArrayManufacture | Represents MAGE Class ArrayManufacture Describes the process by which arrays are produced. |
| ArrayManufactureDeviation | Represents MAGE Class ArrayManufactureDeviation Stores information of the potential difference between an array design and arrays that have been manufactured using that design (e.g. a tip failed to print several spots). |
| ArrayManufactureWIDArrayWID | Represents n..n association between ArrayManufacture and Array Association between the manufactured array and the information on that manufacture. |
| ArrayManufactureWIDContactWID | Represents n..n association between ArrayManufacture and Contact The person or organization to contact for information concerning the ArrayManufacture. |
| BAssayMappingWIDBAssayMapWID | Represents n..n association between BioAssayMapping and BioAssayMap The maps for the BioAssays. |
| BioAssay | Represents MAGE Class BioAssay An abstract class which represents both physical and computational groupings of arrays and biomaterials. Includes MAGE Class DerivedBioAssay Includes MAGE Class MeasuredBioAssay Includes MAGE Class PhysicalBioAssay |
| BioAssayData | Represents MAGE Class BioAssayData Represents the dataset created when the BioAssays are created. BioAssayData is the entry point to the values. Because the actual values are represented by a different object, BioDataValues, which can be memory intensive, the annotation of the transformation can be gotten separate from the data. Includes MAGE Class DerivedBioAssayData Includes MAGE Class MeasuredBioAssayData |
| BioAssayDataCluster | Represents MAGE Class BioAssayDataCluster A mathematical method of higher level analysis whereby BioAssayData are grouped together into nodes. |
| BioAssayDimension | Represents MAGE Class BioAssayDimension An ordered list of bioAssays. |
| BioAssayDimensioWIDBioAssayWID | Represents n..n association between BioAssayDimension and BioAssay The BioAssays for this Dimension |
| BioAssayMapping | Represents MAGE Class BioAssayMapping Container of the mappings of the input BioAssay dimensions to the output BioAssay dimension. |
| BioAssayMapWIDBioAssayWID | Represents n..n association between BioAssayMap and BioAssay The sources of the BioAssayMap that are used to produce a target DerivedBioAssay. |
| BioAssayTuple | Represents MAGE Class BioAssayTuple Transformed container to specify a BioAssay and the Design Elements and their data for that BioAssay. |
| BioAssayWIDChannelWID | Represents n..n association between BioAssay and Channel Channels can be non-null for all subclasses. For instance, collapsing across replicate features will create a DerivedBioAssay that will potentially reference channels. |
| BioAssayWIDFactorValueWID | Represents n..n association between BioAssay and FactorValue The values that this BioAssay is associated with for the experiment. |
| BioDataValues | Represents MAGE Class BioDataValues The actual values for the BioAssayCube. Includes MAGE Class BioDataCube Includes MAGE Class BioDataTuples |
| BioEvent | Represents MAGE Class BioEvent An abstract class to capture the concept of an event (either in the laboratory or a computational analysis). Includes MAGE Class CompositeCompositeMap Includes MAGE Class FeatureReporterMap Includes MAGE Class ReporterCompositeMap Includes MAGE Class Map Includes MAGE Class BioAssayMap Includes MAGE Class DesignElementMap Includes MAGE Class QuantitationTypeMap Includes MAGE Class Transformation Includes MAGE Class Treatment Includes MAGE Class BioAssayCreation Includes MAGE Class BioAssayTreatment Includes MAGE Class FeatureExtraction Includes MAGE Class Hybridization Includes MAGE Class ImageAcquisition |
| BioMaterialMeasurement | Represents MAGE Class BioMaterialMeasurement A BioMaterialMeasurement is a pairing of a source BioMaterial and an amount (Measurement) of that BioMaterial. |
| BioSource | Defines the biological source of an entry in the Protein,
NucleicAcid or Gene tables ("the object"), as well as other tables
eventually storing experimental data related to e.g., microarray or
proteomic experiments. For example, BioSource might be used to specify
the organism, organ, and tissue in which gene expression experiments
were performed. DEPENDENCIES: 1. As BioSource references Taxon.WID to
specify the species, the NCBI taxonomy dataset must have ALREADY been
loaded prior to populating BioSource. 2. BioSource stores subtype
information in BioSubtype via the BioSourceWIDBioSubtypeWID linking
table
|
| BioSourceWIDBioSubtypeWID | Linking table which links BioSource and BioSubtype |
| BioSourceWIDContactWID | Represents n..n association between BioSource and Contact The BioSource's source is the provider of the biological material (a cell line, strain, etc...). This could be the ATTC (American Tissue Type Collection). |
| BioSourceWIDGeneWID | Associates a gene with those species, strains, tissues, etc, from which it was derived. |
| BioSourceWIDProteinWID | Associates a protein with those species, strains, tissues, etc, from which it was derived. |
| BioSubtype | Describes idiosyncratic sub-species descriptions such as "serotype", "variety", etc. These can be many to one (1 BioSource - many BioSubtypes) linked via BioSourceWIDBioSubtypeWID |
| Channel | Represents MAGE Class Channel A channel represents an independent acquisition scheme for the ImageAcquisition event, typically a wavelength. |
| ChannelWIDCompoundWID | Represents n..n association between Channel and Compound The compound used to label the extract. |
| Chemical | Defines small molecular weight chemical compounds. |
| ChemicalAtom | The tables ChemicalAtom and ChemicalBond define the chemical bond structure of a chemical,
the
charge on the constituent atoms, and encodes a two or three dimensional representation of the structure. It is implicit that ChemicalBonds are symmetric. Atoms that are chiral centers are have a non-zero StereoParity field. Values for this field are defined in the Enumeration table, and are taken from MDL Molfile format defined in http://www.mdli.com/downloads/ctfile/ctfile_subs.html . That specification is confusing because it appears to define two redundant ways (marking atoms and marking bonds) of defining stereo configurations. Our theory is that the redundancy exists to allow different drawings of stereo configurations, and that for simply capturing a configuration, setting the StereoParity field of an atom is sufficient. Also, setting the BondStereo field of a bond could simplify drawing of chemical structures, and can allow different drawings of the same stereo configurations, which can be desirable. But these two fields can be interpreted independently of one another. See the appendix of the MDL specification for details on interpreting stereo configurations. E.g. H2O could be encoded as: ChemicalAtom (wid, 1, 'H', 0) ChemicalAtom (wid, 2, 'O', 0) ChemicalAtom (wid, 3, 'H', 0) ChemicalBond (wid, 1, 2, 1) ChemicalBond (wid, 2, 3, 1) |
| ChemicalBond | The tables ChemicalAtom and ChemicalBond define the chemical bond structure of a chemical, the charge
on
the constituent atoms, and encodes a two or three dimensional representation of the structure. It is implicit that ChemicalBonds are symmetric. BondTypes and BondStereo are defined in the Enumeration table, and are taken from MDL Molfile format defined in http://www.mdli.com/downloads/ctfile/ctfile_subs.html . That specification is confusing. See documentation of ChemicalAtom table for more information. E.g. H2O could be encoded as: ChemicalAtom (wid, 1, 'H', 0) ChemicalAtom (wid, 2, 'O', 0) ChemicalAtom (wid, 3, 'H', 0) ChemicalBond (wid, 1, 2, 1) ChemicalBond (wid, 2, 3, 1) |
| Citation | Defines a literature citation. Typically a citation is associated with one or more Warehouse objects via the CitationWIDOtherWID table. |
| CitationWIDOtherWID | Link from citations to the entities described in that citation. Enables one citation to provide support for multiple entries in the warehouse. |
| CommentTable | This table allows for arbitrary association of (possibly lengthy) comments with any object in the warehouse. See the Description table for a more specific type of comment. |
| ComposGrpWIDComposSequenceWID | Represents n..n association between CompositeGroup and CompositeSequence The compositeSequences that belong to this group. |
| CompositeSeqWIDBioSeqWID | Represents n..n association between CompositeSequence and BioSequence The annotation on the BioSequence this CompositeSequence represents. Typically the sequences will be a Genes, Exons, or SpliceVariants. |
| ComposSeqDimensWIDComposSeqWID | Represents n..n association between CompositeSequenceDimension and CompositeSequence The CompositeSequences for this Dimension. |
| ComposSeqWIDComposComposMapWID | Represents n..n association between CompositeSequence and CompositeCompositeMap A map to the compositeSequences that compose this CompositeSequence. |
| ComposSeqWIDRepoComposMapWID | Represents n..n association between CompositeSequence and ReporterCompositeMap A map to the reporters that compose this CompositeSequence. |
| CompoundMeasurement | Represents MAGE Class CompoundMeasurement A CompoundMeasurement is a pairing of a source Compound and an amount (Measurement) of that Compound. |
| Computation | Defines a parameterized computation that has been performed on objects in the warehouse. TODO: Parameters are specified in the Parameter table. |
| Contact | Represents MAGE Class Contact A contact is either a person or an organization. Includes MAGE Class Organization Includes MAGE Class Person |
| CrossReference | This table is used to define (i) links between databases; (ii) links between objects within the same
dataset.
For case (i): A row in this table defines a link between an object in the warehouse (OtherWID) and an entry in another dataset that may or may not be loaded into the warehouse. If it is not loaded, DatabaseName (and XID, Type, and Version if available) will be nonnull. For case (ii): A row in this table defines a link between an object in the warehouse (OtherWID) and an object in the same dataset for which the dataset specifies an association between them not otherwise represented within the warehouse. Such associations are currently (5/10/04) made between a gene described in one sequence (the root Bioseq) and another GenBank entry (the reference Bisoseq) which is being referenced and describes the gene independently of the first Bioseq. In such a case, CrossReference.OtherWID=Gene.WID. In either case: if CrossWID is nonnull, the object linked to is loaded into the warehouse; if it is null, it may or may not be loaded. Note that if a loader does not know whether a referenced entry will be loaded, it is free to fill in CrossWID at a later time. Restrictions: CrossReference only stores keys that point to uniquely identified objects in the referenced database. |
| Database_ | Represents MAGE Class Database An address to a repository. |
| DatabaseWIDContactWID | Represents n..n association between Database and Contact Information on the contacts for the database |
| DataExternal | Represents MAGE Class DataExternal Transformed class to associate external data to the BioAssayDataCube |
| DataInternal | Represents MAGE Class DataInternal Transformed class to associate whitespaced delimited data to the BioAssayDataCube |
| DataSet | Table whose rows define each dataset currently loaded into the warehouse. |
| DataSetHierarchy | Defines a dataset containment hierarchy.
If a dataset contains another, either directly or indirectly,
a row exists for that dataset pair in this table.
A dataset always contains itself; therefore each dataset is included in at least one row in this table.
A dataset may contain any number of datasets, and be contained in any number of datasets.
To query for an object in a containing dataset whose datasetwid is NN,
regardless of which contained dataset it is in, use a query like the following: select wid from DataSetHierarchy h, Protein p where h.superwid=NN and h.subwid=p.datasetwid and p.name='some name' |
| Datum | Represents MAGE Class Datum Transformed container to hold a value. QuantitationType will determine the type of this value. |
| DBID | Associates a warehouse entity with the identifier(s) used for that entity in its source dataset. For example, if a protein is loaded from SwissProt into the warehouse, this table can be used to store the SwissProt accession numbers for the protein. Restrictions: DBID only stores keys that point to uniquely identified objects in the referenced database. |
| DerivBioAssayWIDBioAssayMapWID | Represents n..n association between DerivedBioAssay and BioAssayMap The DerivedBioAssay that is produced by the sources of the BioAssayMap. |
| DerivBioAWIDDerivBioADataWID | Represents n..n association between DerivedBioAssay and DerivedBioAssayData The data associated with the DerivedBioAssay. |
| Description | This table contains a textual description of a warehouse object. An object will not have more than one description, and the description text will typically define or otherwise characterize the object. See the CommentTable table for a more general type of comment. |
| DesignElement | Represents MAGE Class DesignElement An element of an array. This is generally of type feature but can be specified as reporters or compositeSequence for arrays that are abstracted from a physical array. Includes MAGE Class CompositeSequence Includes MAGE Class Feature Includes MAGE Class Reporter |
| DesignElementDimension | Represents MAGE Class DesignElementDimension An ordered list of designElements. It will be realized as one of its three subclasses. Includes MAGE Class CompositeSequenceDimension Includes MAGE Class FeatureDimension Includes MAGE Class ReporterDimension |
| DesignElementGroup | Represents MAGE Class DesignElementGroup The DesignElementGroup holds information on either features, reporters, or compositeSequences, particularly that information that is common between all of the DesignElements contained. Includes MAGE Class CompositeGroup Includes MAGE Class FeatureGroup Includes MAGE Class ReporterGroup |
| DesignElementMapping | Represents MAGE Class DesignElementMapping Container of the mappings of the input DesignElement dimensions to the output DesignElement dimension. |
| DesignElementTuple | Represents MAGE Class DesignElementTuple Transformed container to specify a DesignElement and QuantitationTypes for that Element. |
| DesnElMappingWIDDesnElMapWID | Represents n..n association between DesignElementMapping and DesignElementMap The maps for the DesignElements. |
| Division | Stores the GenBank Division and the NCBI three letter code for records originating from GenBank.
NCBI uses Divisions to organize GenBank records according to two definitions: some divisions are based on
taxonomy (e.g., the "BCT" division), whereas other divisions exist purely for data organization reasons
(e.g., the "CON" division, whose records store instructions for the construction of large contigs).
Sequences from different GenBank divisions vary substantially in quality based upon the way they were
generated, e.g., single-pass genomic sequencing, ESTs (low quality, single-pass sequencing), cDNAs
(high coverage sequencing) and genomic sequencing (moderate coverage sequencing). All BioWarehouse
records associated with a GenBank dataset also have a record in the Division table.
|
| Element | The periodic table of elements |
| Entry | Defines metadata describing a warehouse object. Every warehouse object (that is, anything with a WID column) must have an associated Entry row. |
| Enumeration | This table defines enumerated types used within the warehouse. Essentially, enumerated types correspond to small controlled vocabularies used within one or more warehouse tables. For example, the warehouse table Feature has a column called Type that defines the type of a feature within an amino-acid sequence. Examples types might be PHOS-RESIDUE and GLY-RESIDUE, corresponding to residues in a protein that are phosphorylated and glycosylated, respectively. Those two enumerated types would be entered into two rows of this Enumeration table. Note that in some cases the values actually stored in the warehouse table are numbers, which are stored in the Value field of the Enumeration table as the string form of that number. |
| EnzReactionAltCompound | Identifies a compound that is either an alternate substrate or an alternate cofactor to a primary compound present in an enzymatic reaction. |
| EnzReactionCofactor | Cofactors are chemicals that are required for the enzyme to catalyze the reaction, but are left unchanged by the reaction. If multiple cofactors are listed for a reaction, this is interpreted as a disjunction. This table also encodes prosthetic groups. |
| EnzReactionInhibitorActivator | Associates an enzymatic reaction to the compounds that act as inhibitors and activators for the reaction. The mechanism of action is encoded in the Mechanism column. |
| EnzymaticReaction | Defines an association between a reaction and an enzyme that catalyzes that reaction (ProteinWID). In the case where we are defining an association between a subunit of a larger enzyme complex, and a reaction catalyzed by that subunit only when the subunit is part of that larger complex, ComplexWID specifies that larger complex (assumes that complex is in the warehouse). This table requires a WID as it is referenced in multiple linking tables to associate alternate compounds, cofactors, etc., to allow for multiple valued attributes of the enzymatic reaction. |
| Experiment | Defines an experiment. Provides a context for associating experimental data with it. Allows tree-structured experiments consisting of heterogeneous subexperiments, subexperiments corresponding to time-series observations, and repeated trials of identical experiments. For a hierarchical experiment, data should be associated with the Experiment at the appropriate level. For example, if data reflects results from averaging numerous identically conducted trials, that data should be associated with the Experiment representing the group of these trials. A Comment table entry may be created to contain discussion of results, etc. A DBID table entry may be created if the (sub)experiment has a unique name within its DataSet. SynonymTable table entries may be created to associate names with the (sub)experiment. If published, a reference may be created in Citation table. |
| ExperimentalFactor | Represents MAGE Class ExperimentalFactor ExperimentFactors are the dependent variables of an experiment (e.g. time, glucose concentration, ...). |
| ExperimentData | Specifies a relationship between one data entity and an experiment in which it was
recorded as an observation or used in some other fashion. Used in flow cytometry to represent both observations and filter wavelengths. For an observation: ExperimentData.Data contains the vector of observations ExperimentData.MageData = NULL ExperimentData.OtherWID = the FlowCytometrySample.WID of the sample, ExperimentData.Role defines the reading ('forward scatter', 'side scatter', or 'filter'), ExperimentData.Type = 'flow cytometry', ExperimentData.Kind = 'O' (observation) For a filter wavelength: ExperimentData.Data = NULL ExperimentData.MageData references a ParameterValue row for the wavelength ExperimentData.Role = 'filter wavelength' ExperimentData.Kind = 'P' (parameter) ExperimentData.OtherWID = the ExperimentData.WID of the observation, |
| ExperimentDesign | Represents MAGE Class ExperimentDesign The ExperimentDesign is the description and collection of ExperimentalFactors and the hierarchy of BioAssays to which they pertain. |
| ExperimentDesignWIDBioAssayWID | Represents n..n association between ExperimentDesign and BioAssay The organization of the BioAssays as specified by the ExperimentDesign (TimeCourse, Dosage, etc.) |
| ExperimentRelationship | Relates one experiment to another. This table supports a many to many relationship. |
| ExperimentWIDBioAssayDataWID | Represents n..n association between Experiment and BioAssayData The collection of BioAssayDatas for this Experiment. |
| ExperimentWIDBioAssayWID | Represents n..n association between Experiment and BioAssay The collection of BioAssays for this Experiment. |
| ExperimentWIDContactWID | Represents n..n association between Experiment and Contact The providers of the Experiment, its data and annotation. |
| ExperimWIDBioAssayDataClustWID | Represents n..n association between Experiment and BioAssayDataCluster The results of analyzing the data, typically with a clustering algorithm. |
| FactorValue | Represents MAGE Class FactorValue The value for a ExperimentalFactor |
| Feature | Features define regions or points of interest on the protein sequence or nucleic acid sequence specified by SequenceWID. Features are not used to describe objects such as Genes, BioSources, etc, because attributes of those objects are described in their own tables. Exceptions: 1. Since pseudogenes are not entered in the Gene table, they are listed as sequence features in Feature |
| FeatureDefect | Represents MAGE Class FeatureDefect Stores the defect information for a feature. |
| FeatureDimensionWIDFeatureWID | Represents n..n association between FeatureDimension and Feature The features for this dimension. |
| FeatureInformation | Represents MAGE Class FeatureInformation As part of the map information, allows the association of one or more differences in the BioMaterial on a feature from the BioMaterial of the Reporter. Useful for control purposes such as in Affymetrix probe pairs. |
| FeatureLocation | Represents MAGE Class FeatureLocation Specifies where a feature is located relative to a grid. |
| FeatureWIDFeatureWID | Represents n..n association between Feature and Feature Associates features with their control features. |
| FeatureWIDFeatureWID2 | Represents n..n association between Feature and Feature Associates features with their control features. |
| Fiducial | Represents MAGE Class Fiducial A marking on the surface of the array that can be used to identify the array's origin, the coordinates of which are the fiducial's centroid. |
| FlowCytometryProbe | Defines a material used to measure characteristics of a cell using flow cytometry.
If probe is a protein, a CrossReference may be defined to specify the protein;
in that case CrossReference.OtherWID refers to this FlowCytometryProbe.
|
| FlowCytometrySample | Defines an experimental sample prepared for use in a flow cytometry experiment.
A typical experiment consists of multiple samples X multiple readings
(forward/side scatter and using filters of various wavelengths),
with a vector of observations (one per cell of the BioSource)
for each sample/reading combination. An ExperimentData row is defined for each observation; see that table for representation details. An Experiment row is defined to associate each ExperimentData row with the experiment. |
| Function | Describes the non-enzymatic function(s) of proteins. That is, this table should not be used to describe the enzymatic function of a protein whose enzymatic function is described using the EnzymaticReaction table. Function names are stored as strings with no particular format or interpretation. |
| GelLocation | Describes the location of a spot on a gel. This tables maps spots to the one or more tables that they are found on. Describes the X and Y coordinates on the Gel. Information for the particular gel conditions that the spot was identified on can be found in the Experiment table. |
| Gene | Defines a notion of gene that is limited to procaryotic aspects of genes. Later versions of the warehouse will expand this definition to include eukaryotic aspects of genes. Separate tables define associations between a gene and (1) its biological source(s), (2) its protein product(s), if any, and (3) its RNA product(s), if any. |
| GeneExpressionData | MAGE gene expression data (BioDataCube) This table definition may evolve as the requirements for gene expression data are better understood. The MAGE OM currently allows 'any' type for the Value component of the BioDataCube. We are investigating ways to make this table better accomodate the variety of allowed datatypes. Please send suggestions to support@biowarehouse.org |
| GeneticCode | Defines the genetic codes with a name, translation table and a start codon for each code. See http://embryology.med.unsw.edu.au/DNA/Genetic_Codes.htm for more information. Genbank ID is stored as DBID.XID, where DBID.OtherWID = GeneticCode.WID. |
| GeneWIDNucleicAcidWID | Associates a gene with its nucleic acid product(s). Note that the nucleic acid (ie. replicon) containing the gene is referenced by Gene.NucleicAcidWID, not in this table. |
| GeneWIDProteinWID | Associates a gene with its protein product(s). |
| HardwareWIDContactWID | Represents n..n association between Hardware and Contact Contact for information on the Hardware. |
| HardwareWIDSoftwareWID | Represents n..n association between Hardware and Software Associates Hardware and Software together. |
| Image | Represents MAGE Class Image An image is created by an imageAcquisition event, typically by scanning the hybridized array (the PhysicalBioAssay). |
| ImageAcquisitionWIDImageWID | Represents n..n association between ImageAcquisition and Image The images produced by the ImageAcquisition event. |
| ImageWIDChannelWID | Represents n..n association between Image and Channel The channels captured in this image. |
| Interaction | Defines a molecular interaction, such as two proteins observed to interact in a yeast-two-hybrid
experiement. The interaction could involve macromolecules, small molecules or both.
|
| InteractionParticipant | Associates an interaction with the entity that participates in the interaction. |
| LabeledExtractWIDCompoundWID | Represents n..n association between LabeledExtract and Compound Compound used to label the extract. |
| LightSource | Defines a light source, e.g., as produced by a scientific instrument.
Used by flow cytometry in describing the cytometry instrument.
|
| Location | Defines one or more locations of a protein. |
| ManufactureLIMS | Represents MAGE Class ManufactureLIMS Information on the physical production of arrays within the laboratory. Includes MAGE Class ManufactureLIMSBiomaterial |
| MeasBAssayWIDMeasBAssayDataWID | Represents n..n association between MeasuredBioAssay and MeasuredBioAssayData The data associated with the MeasuredBioAssay. |
| Measurement | Represents MAGE Class Measurement A Measurement is a quantity with a unit. |
| MismatchInformation | Represents MAGE Class MismatchInformation Describes how a reporter varies from its ReporterCharacteristics sequence(s) or how a Feature varies from its Reporter sequence. |
| NameValueType | Represents MAGE Class NameValueType A tuple designed to store data, keyed by a name and type. |
| Node | Represents MAGE Class Node An individual component of a clustering. May contain other nodes. |
| NodeContents | Represents MAGE Class NodeContents The contents of a node for any or all of the three Dimensions. If a node only contained genes just the DesignElementDimension would be defined. |
| NodeValue | Represents MAGE Class NodeValue A value associated with the Node that can rank it in relation to the other nodes produced by the clustering algorithm. |
| NucleicAcid | Defines a specific nucleic acid molecule, such as DNA or of RNA. Entries in this table correspond to a real-world DNA or RNA molecules which at one point were purified and isolated ("something you can point at"). A row in this table can define a complete molecule, a fragment of a molecule, or a molecule that has been partially sequenced in different regions. This table will be used in several ways: (1) to associate a sequence with an entire replicon, or a region of a replicon, when the sequence of that replicon is known; (2) to associate a DNA sequence with a single gene; (3) to define a DNA or RNA molecule that has not been sequenced; (4) to define RNA molecules such as tRNAs. Features on a NucleicAcid molecule (such as promoters or binding sites) can be defined using the Feature table. The Subsequence table contains zero or more full or partial sequences contents of this molecule. If one represents the full sequence of this molecule, Subsequence.FullSequence = 'T' where Subsequence.NucleicAcidWID references this molecule. In this case, NucleicAcid.FullySequenced = 'T' as well. |
| Parameter | Represents MAGE Class Parameter A Parameter is a replaceable value in a Parameterizable class. Examples of Parameters include: scanning wavelength, laser power, centrifuge speed, multiplicative errors, the number of input nodes to a SOM, and PCR temperatures. |
| Parameterizable | Represents MAGE Class Parameterizable The Parameterizable interface encapsulates the association of Parameters with ParameterValues. Includes MAGE Class Hardware Includes MAGE Class Protocol Includes MAGE Class Software |
| ParameterizableApplication | Represents MAGE Class ParameterizableApplication The interface that is the use of a Parameterizable class. Includes MAGE Class HardwareApplication Includes MAGE Class ProtocolApplication Includes MAGE Class SoftwareApplication |
| ParameterValue | Represents MAGE Class ParameterValue The value of a Parameter. |
| Pathway | Pathways are graphs of reactions, grouped together according to a higher biological function they perform. Some pathways may be "template", "reference", "model" or "sum of organisms" pathways. Those pathways do not contain an BioSourceWID reference, and have the type field set to "R" (Reference/Model/Theoretical). For real organisms, the type is "O". |
| PathwayLink | This table allows us to state that two pathways are neighbors in a biologically significant sense, because they share a substrate in common. This link between two pathways is represented by the WIDs of the interacting pathways, and of the chemical compound that they share. |
| PathwayReaction | A pathway is defined as a set of reaction pairs or a collection of molecular interactions between
molecules.
This table stores not only the relationships between pathway and reactions, but also relationships between
molecular interaction network/pathway and interactions.
For each pair of reactions R1 and R2, R1 directly precedes R2 in the pathway.
Some reactions in a pathway may be considered hypothetical, probably because the presence of the enzyme
has not been demonstrated.
To specify an interaction is a part of a molecular interaction network/pathway, use R2 to represents
a molecular interaction, leave R1 blank.
|
| Position_ | Represents MAGE Class Position Specifies a position on an array. |
| PositionDelta | Represents MAGE Class PositionDelta The delta the feature was actually printed on the array from the position specified for the feature in the array design. |
| Product | Associates a reaction with a chemical product of that reaction. |
| Protein | Defines a specific protein, that is, a real-world protein which was either purified and
isolated,
OR is reasonably inferred by genomic analysis or other means, such as enzymological characterization. The protein could be a monomer or a multimer. In the latter case, a sequence would not be stored for such a record. |
| ProteinWIDFunctionWID | Associates a non-enzymatic function with a protein that has that function. |
| ProteinWIDSpotWID | Associates a spot with a known protein. Not all spots are associated with proteins since it is not necessarily known what the protein is for a given spot. This table supports the ability to have a spot linked to multiple protein, or a protein linked to multiple spots. |
| ProtocolApplWIDPersonWID | Represents n..n association between ProtocolApplication and Person The people who performed the protocol. |
| ProtocolWIDHardwareWID | Represents n..n association between Protocol and Hardware Hardware used by this protocol. |
| ProtocolWIDSoftwareWID | Represents n..n association between Protocol and Software Software used by this Protocol. |
| QuantitationType | Represents MAGE Class QuantitationType A method for calculating a single datum of the matrix (e.g. raw intensity, background, error). Includes MAGE Class SpecializedQuantitationType Includes MAGE Class StandardQuantitationType Includes MAGE Class ConfidenceIndicator Includes MAGE Class DerivedSignal Includes MAGE Class Error Includes MAGE Class ExpectedValue Includes MAGE Class Failed Includes MAGE Class MeasuredSignal Includes MAGE Class PValue Includes MAGE Class PresentAbsent Includes MAGE Class Ratio |
| QuantitationTypeDimension | Represents MAGE Class QuantitationTypeDimension An ordered list of quantitationTypes. |
| QuantitationTypeMapping | Represents MAGE Class QuantitationTypeMapping Container of the mappings of the input QuantitationType dimensions to the output QuantitationType dimension. |
| QuantitationTypeTuple | Represents MAGE Class QuantitationTypeTuple Transformed container to specify a Quantitation Type and the value for that Type. |
| QuantTyMapWIDQuantTyMapWI | Represents n..n association between QuantitationTypeMapping and QuantitationTypeMap The maps for the QuantitationTypes. |
| QuantTypeDimensWIDQuantTypeWID | Represents n..n association between QuantitationTypeDimension and QuantitationType The QuantitationTypes for this Dimension. |
| QuantTypeMapWIDQuantTypeWID | Represents n..n association between QuantitationTypeMap and QuantitationType The QuantitationType sources for values for the transformation. |
| QuantTypeWIDConfidenceIndWID | Represents n..n association between QuantitationType and ConfidenceIndicator The association between a ConfidenceIndicator and the QuantitationType its is an indicator for. |
| QuantTypeWIDQuantTypeMapWID | Represents n..n association between QuantitationType and QuantitationTypeMap The QuantitationType whose value will be produced from the values of the source QuantitationType according to the Protocol. |
| Reactant | Associates a reaction with a chemical that is consumed by the reaction. |
| Reaction | Defines a chemical reaction. The reaction could be enzyme catalyzed or occur spontaneously. The reaction could involve small molecules, macromolecules, or a combination of the two. Every reaction will be stored in the warehouse in a given direction, for example, every reaction that has an assigned EC number is written in a direction assigned by the enzyme commission. In physiological settings, the reaction could occur in the direction the reaction is stored, the reverse direction, or both directions. Restrictions: Reaction only stores fully qualified EC numbers, e.g., NOT of the form "X.Y.Z.-" |
| RelatedTerm | Defines a relationship between a term and another object.
For relationships between terms, TermRelationship is generally used.
|
| ReporterDimensWIDReporterWID | Represents n..n association between ReporterDimension and Reporter The reporters for this dimension. |
| ReporterGroupWIDReporterWID | Represents n..n association between ReporterGroup and Reporter The reporters that belong to this group. |
| ReporterWIDBioSequenceWID | Represents n..n association between Reporter and BioSequence The sequence annotation on the BioMaterial this reporter represents. Typically the sequences will be an Oligo Sequence, Clone or PCR Primer. |
| ReporterWIDFeatureReporMapWID | Represents n..n association between Reporter and FeatureReporterMap Associates features with their reporter. |
| SeqFeatureLocation | Represents MAGE Class SeqFeatureLocation The location of the SeqFeature annotation. |
| SequenceMatch | Records a result of a computation of the degree of match between two sequences. Sequences are either both Proteins or both Subsequences. |
| SequencePosition | Represents MAGE Class SequencePosition Designates the position of the Feature in its BioSequence. Includes MAGE Class CompositePosition Includes MAGE Class ReporterPosition |
| SoftwareWIDContactWID | Represents n..n association between Software and Contact Contact for information on the software. |
| SoftwareWIDSoftwareWID | Represents n..n association between Software and Software Software packages this software uses, i.e. operating system, 3rd party software packages, etc. |
| SpecialWIDTable | Special WIDs are low-valued WIDs, assigned to globally and frequently used WIDs within a dataset, such as its DataSetWID. They are both more mnemonic, and potentially more efficient to store. Warehouse.MaxSpecialWID delimits special and regular WIDs. Analogous to WIDTable, this table is used to allocate special WIDs. MySQL dataset loaders use the following protocol for WID assignment: DELETE FROM SpecialWIDTable; INSERT INTO SpecialWIDTable VALUES (); SELECT last_insert_id(); This table contains exactly one row at any given time. |
| Spot | Describes a spot from a 2D Gel experiment. A spot can be linked to one or more gels and the location on the gel. The gel location information is stored in the ExperimentData table. A spot can be identified to be a particluar protein in the protein table and is associated to proteins via the ProteinWIDSpotWID table The method used to identify the spot is contained in the SpotWIDSpotIDMethodWID table and the SpotIDMethod table. The growth conditions for the protein is found in the ExperimentData tableeh The Spot Name/ID goes into the SpotId row, but additional names can be stored as DBID table entry. |
| SpotIdMethod | Describes the method by which a spot was identified as a particular (known) protein in the protein table. |
| SpotWIDSpotIdMethodWID | Associates a spot with a SpotIdMethod, which is the method used to identify a spot with a particular protein. A spot can be identified as a particular protein by multiple methods, so this table supports this many-to-many relationship. |
| Subsequence | Contains a contiguous sequence of a nucleic acid molecule. Multiple Subsequences can make up a given
NucleicAcid entry.
Contains either the full or partial nucleotide sequence of the nucleic acid molecule stored in NucleicAcid. Todo: Currently the schema has no way of indicating an approximate position of this Subsequence on the NucleicAcid as a whole. |
| Subunit | Specifies that Subunit is a subunit of Complex. These subunit relationships could be described in multiple levels. For example, this table can be used to describe multimeric proteins, or ribosomes. |
| SuperPathway | Pathways may be arranged in a hierarchy, i.e. containment within one or more superpathways, as an abstraction mechanism. |
| Support | Describes the source and strength of evidence for individual facts in the warehouse. This table is designed to allow different types of evidential support to be asserted for different types of database assertions, such as using one set of evidence types for supporting assertions about protein function, and another, possibly overlapping set of evidence types for supporting assertions about protein existence. Example use: Record that there are two sources of support for the function of a given protein, one computational and one experimental. This table has a WID for use in the CitationWIDOtherWID table. |
| SynonymTable | Defines one or more synonyms for a warehouse entity such as a protein, a gene, a small molecule, or a pathway. |
| Taxon | Defines a taxon, which could be a species, genus, family or some other rank. These ranks are controlled vocabulary and are stored in the Enumeration table. The taxon table associates a taxon with its parent, gives the name, division code and the genetic code information for the taxon. |
| Term | A term is a controlled vocabulary term used within a particular dataset. E.g. the keyword attached to proteins in SwissProt. Terms can be arranged in a hierarchy (see the TermRelationship table). |
| TermRelationship | Defines a relationship between two terms. |
| ToolAdvice | This table captures meta-data that does not have an interpretation in the warehouse semantics, but is `hints' to various tools that may operate on, or build pictorial representations of, core elements of the schema. For example, it may give hints to a particular pathway viewing tool on how to layout a pathway. |
| TranscriptionUnit | Defines a transcription unit, similar to an operon. There is a one-to-one
relationship between a promoter and a transcription unit. The transcription
unit includes the promoter, nearby site that regulate its activity, and downstream
genes and the terminator. Unlike operons, a transcription unit can contain a single
gene, and a transcription unit can contain only one promoter. For operons containing
multiple promoters, a different transcription unit would be created for each operon.
See TranscriptionUnitComponent for the types of components represented.
|
| TranscriptionUnitComponent | Associates a TranscriptionUnit with its components. Components consist of
genes and/or features. A feature component may be either a promoter, a terminator or a DNA binding site
(transcription-factor binding site).
|
| TransformWIDBioAssayDataWID | Represents n..n association between Transformation and BioAssayData The BioAssayData sources that the Transformation event uses to produce the target DerivedBioAssayData. |
| Unit | Represents MAGE Class Unit The unit is a strict enumeration of types. Includes MAGE Class ConcentrationUnit Includes MAGE Class DistanceUnit Includes MAGE Class MassUnit Includes MAGE Class QuantityUnit Includes MAGE Class TemperatureUnit Includes MAGE Class TimeUnit Includes MAGE Class VolumeUnit |
| Valence | Defines one or more valences for each element. |
| Warehouse | Describes this version of the BioSPICE Warehouse. This table contains exactly one row. |
| WIDTable | To allow for compatibility with RDBMS systems that do not support sequences, this table may be used to store the last used WID. MySQL dataset loaders use the following SQL for WID assignment: DELETE FROM WIDTable; INSERT INTO WIDTable VALUES (); SELECT last_insert_id(); It may also be used in RDBMS systems that do not support auto-increment of a counter; however additional protocol like table locking may be needed to assure the integrity of WIDs during concurrent assignment. This table contains exactly one row at any given time. |
| Zone | Represents MAGE Class Zone Specifies the location of a zone on an array. |
| ZoneDefect | Represents MAGE Class ZoneDefect Stores the defect information for a zone. |
| ZoneGroup | Represents MAGE Class ZoneGroup Specifies a repeating area on an array. This is useful for printing when the same pattern is repeated in a regular fashion. |
| ZoneLayout | Represents MAGE Class ZoneLayout Specifies the layout of features in a rectangular grid. |